Collaborative RMarkdown Hack
Document editing and report creation are key parts of any researcher’s workflow.
Thankfully, the tools to edit research documents have improved greatly over the last few years. While I used to write most documents with $ \LaTeX $, I’d now recommend that researchers use R Markdown.
R Markdown allows the use of R code in a Markdown format and is also easily extended with features. For example, there is the excellent bookdown package which allows easy use of book formatting, bibliographies and cross-references with Markdown.
It’s very useful when editing an R Markdown document, to be able to share that document with other users. One way to do this is by using CoCalc, which can also be self-hosted. However, the interface for CoCalc can be a bit complicated for first time users. We especially want paper reviewers and editors to find the interface easy to use.
My favourite open-source Markdown editor is CodiMD (soon to be renamed to HedgeDoc). It can be easily installed on a Linux server and allows real-time collaborative editing of Markdown. However… it doesn’t process R Markdown.
So we have three programs that don’t quite cover our use case of a simple, multi-user capable R Markdown editor:
| application | supports R Markdown | collaborative editing | beginner friendly |
|---|---|---|---|
| CoCalc | Yes | Yes | No |
| RStudio | Yes | No | No |
| CodiMD (HedgeDoc) | normal Markdown only | Yes | Yes |
So, how can we solve this problem?
Like we solve most problems in open-source software, combining two tools to make a new one.
In this case, we’ll use the viewer in RStudio to bring the Markdown document from CodiMD.
Then, we can edit the document in CodiMD (and share it with other editors) and use RStudio when we want to render the finished document.
we can also script the rendering of the document at periodic intervals to provide a live document, but that’s a hack for another day
Example integration of RStudio and CodiMD:
- RStudio domain: https://r.example.net
- CodiMD domain: https://codimd.example.net
install an RStudio Server (open source) and CodiMD server (open source)
set up cross site permissions in the web server (or else we can’t embed the viewer)
Header set Content-Security-Policy: "frame-ancestors 'self' https://r.example.net"- create an R project to display editor and pull markdown from CodiMD
setwd("~/R Projects/RMarkdown_Test/")
# load CodiMD into the viewer
# more info on the viewer here: https://support.rstudio.com/hc/en-us/articles/202133558-Extending-RStudio-with-the-Viewer-Pane
viewer <- getOption("viewer")
viewer("https://codimd.example.net/RMarkdown_Test?both")
# Download working file from hackmd and save the file locally
download.file(url='https://codimd.example.net/RMarkdown_Test/download', method='wget', destfile='RMarkdown_Test.Rmd' )
# - now process your R-markdown however you like in RStudioThen, we get something like this:
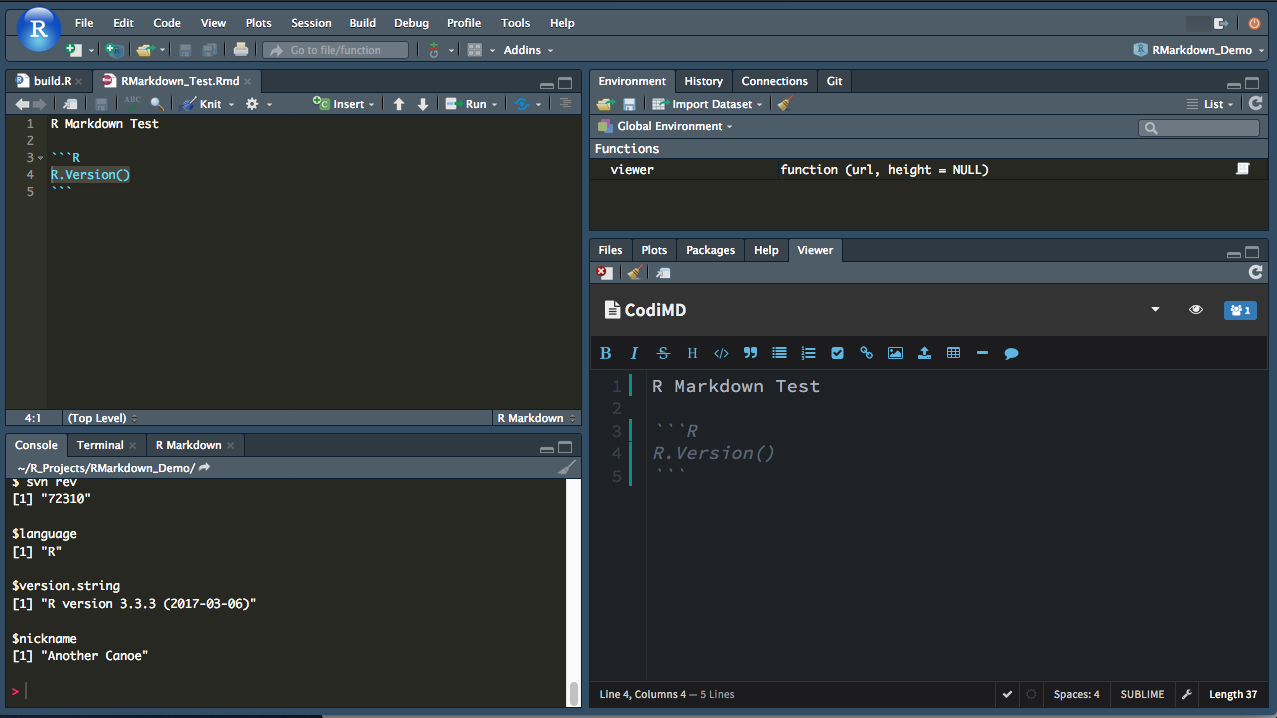
Running the R script now pulls the markdown from CodiMD and can render it to RMarkdown, or bookdown etc.
